TEES is available as a stable version (recommended, downloadable from the above icons) or the latest development version. To get started, read the Quick Start guide.
For documentation, see the Wiki and the API docs.
Current
- Convolutional Neural Network models for TEES
- TEES is used in the EPE 2018 shared task
Overview
Turku Event Extraction System (TEES) is a free and open source natural language processing system developed for the extraction of events and relations from biomedical text. It is written mostly in Python, and should work in generic Unix/Linux environments.
Biomedical event extraction refers to the automatic detection of molecular interactions from research articles. Events can accurately represent complex statements, e.g. the sentence “Protein A causes protein B to bind protein C” produces the event CAUSE(A, BIND(B, C)). Such formal structures can be processed with computational methods, allowing large-scale analysis of the literature, as well as applications such as pathway construction.
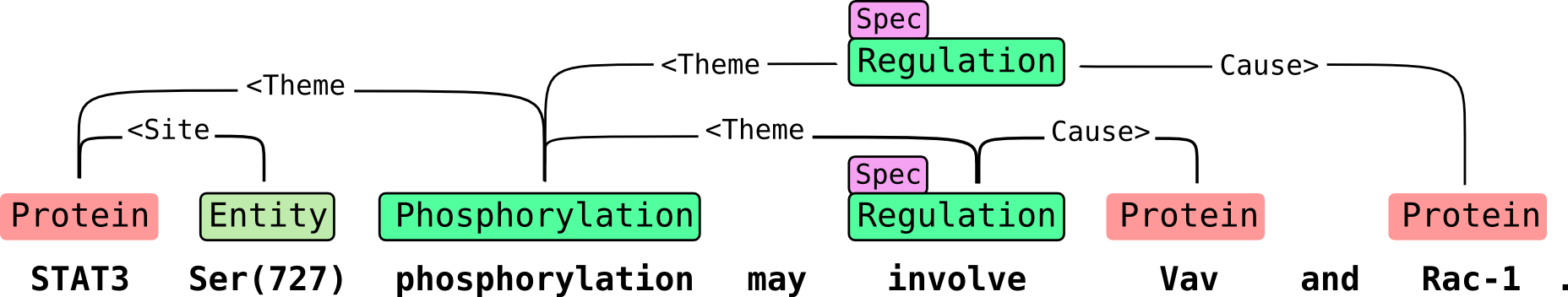
TEES has been evaluated in the following Shared Tasks. Models for predicting their targets are provided with TEES and can be used on any unannotated text.
- BioCreative VI Track 5: CHEMPROT 2017 (Chemical-Protein interactions) (3rd place, PDF)
- DDI 2013 (Drug-drug interactions) Challenge (2nd and 3rd place in the DDI and Drug-NER tasks, PDF)
- BioNLP 2013 Shared Task (1st place in 4/8 tasks, PDF)
- DDI 2011 (Drug-drug interactions) Challenge (4th place, at 96% of the performance of the best system, PDF)
- BioNLP 2011 Shared Task (1st place in 4/8 tasks, only system to participate in all tasks, PDF)
- BioNLP 2009 Shared Task (1st place, PDF)
For a list of all scientific publications on TEES, see the Publications page. TEES has been used to successfully predict GENIA type events for all PubMed abstracts and PMC full text articles, and the resulting data is available in the EVEX database.